Translate this page into:
The use of mitochondrial DNA and short tandem repeat typing in the identification of air crash victims
The identification of dead bodies in mass casualties has been a Herculean task since time immemorial. Fatal crash of civil or military airliners generally leaves numerous unidentified bodies. As compared to other mass casualties viz. earthquakes, floods and collapse of building, the identification of bodies is more difficult post-aircraft accidents as the bodies are mutilated to a large extent and at times decomposed when the aircraft crashes in remote places and recovery is delayed [1]. There have been numerous modalities in identifying the dead in mass casualties like sex, body stature, identification by next of kin, face (if not mutilated), medals (in case worn by the military pilots), identification discs worn by the pilots, personal metal belongings e.g. metal chain, rings and dentition.
The latest modality in identification has been DNA extraction and fingerprinting, which identifies each and every body impeccably. There are various techniques using DNA, which aids in identification of dead in a fatal air crash [2].
Historical aspects
The first wide-scale use of DNA testing was not in medical diagnostics but in forensic application. DNA testing dates back to 1985. Ludes B et al did a medico-legal investigation and identification after the air crash of the Airbus A-320 upon the Mount Sainte-Odile (France). The identification team rapidly retrieved and identified 85 of the 87 victims, with 17 being identified only by DNA typing [3].
Advantages of DNA over traditional methods
The first major advantage is its application to all biological source material. The second major advantage is that DNA testing has tremendous discriminatory potential and a very high sensitivity. The third advantage is its resistance to environmental factors. DNA is a robust molecule resistant to acids, alkalis, detergents, oil, gasoline, high temperatures etc. [4].
Impact of DNA
Smith A et al. studied the success of disaster victim identification in military aviation accident fatalities in the Australian Defence Force (ADF) [5]. The autopsy reports for 117 fatalities were reviewed (covering 73.7% of aircrew fatalities from 1960-2002). Of the 117 victims, 38 (32.4%) sustained injuries, which were severe enough to prevent identification by traditional (non-DNA) techniques viz. fingerprint and dental analysis. These cases were positively identified through the use of DNA techniques [5]. The common availability of families as source of reference material for comparison purpose in DNA typing overcomes the great impediment faced in fingerprint and dental identification.
DNA degradation and environmental damage
DNA normally undergoes progressive fragmentation or degradation reducing the high molecular weight DNA to low molecular weight DNA; however sequence information is still present. Smaller DNA fragments are present for considerable period and allow for some DNA testing.
DNA polymorphism
The DNA from every human except identical twins is unique because of presence of polymorphism. On an average, only one in thousand base pairs differs which explains why all human beings have two legs, two hands, one nose etc. But in three million base pair sequence, there are quite a few differences. These polymorphisms are divided into two types (i) Length polymorphism and (ii) Sequence polymorphism.
Length based polymorphism are found in repetitive DNA. More than 90% of our DNA is superfluous. Approximately 20-30% of non-coding regions are composed of repetitive regions. Variable non-tandem repeat loci in DNA possess a core sequence that is repeated in a string form. The number of core repeats varies from individual to individual.
Sequence polymorphism is composed of different nucleotides at a particular location in the genome. These sequenced variations can be manifested as base substitutions, additions or deletions.
DNA typing methods
Two basic procedures are used- (i) Restriction Fragment Length Polymorphism (RFLP) testing (ii) Polymerase Chain Reaction (PCR) based systems
RFLP
Examination of DNA from any two persons reveals variations in the DNA sequences involving approximately a nucleotide in 200 to 500 base pair stretches. Most of these variations occur in noncoding regions of the DNA and hence are phenotypically silent; however these single base pair changes may abolish or create recognition sites for restriction enzymes, thereby altering the length of DNA fragment produced after digestion with certain restriction enzymes. Using appropriate DNA probes that hybridize with sequences in the vicinity of the polymorphism site, it is possible to detect the DNA fragments of different lengths by Southern Blot analysis. The potential pitfalls of RFLP are that it requires 4-6 weeks time for analysis. Even a slight technical error leads to a false result. Some factors that could produce misleading results are partial degradation of the DNA, which can lead to band shifting [6].
PCR based system
PCR is a technical breakthrough that has revolutionized DNA testing and indeed, the biological sciences. All current non RFLP DNA tests are PCR based methods including (i) Dotblots (ii) Amplified DNA fragment polymorphism and short tandem repeats and (iii) Mitochondrial DNA sequencing. The main advantages of PCR are (i) It is a faster method (ii) It produces more discrete results (iii) It can be performed on cadaveric tissue, form din fixed tissue, and on blood that has been exposed to environment for long.
PCR -Dotblot
Dotblots involve a series of DNA probes to detect target sequences. In traditional dotblot formats, a portion of amplified DNA (from a thermocycler) is bound to a membrane and probed by an SSO probe, any unbound probe is washed away, and a positive test result is visualized by color reaction with the enzyme linked to probe. The commercial kits use a reverse dotblot in which the probe is bound to the membrane and amplified DNA is added to it.
Amplified DNA Fragment Polymorphism and Short Tandem Repeats
AmpFLP analysis involves the electrophoretic separation achieved by size of polymorphic DNA fragments produced by amplification rather then excision, as in traditional RFLP. It basically differentiates on basis of length polymorphism. Variable Number Tandem Repeat (VNTR) loci used are much smaller than RFLP loci as PCR does not reliably amplify large stretches of DNA. AmpFLP electrophoresis is carried out in polyacrylamide gel, which offers higher resolution. There is no background in AmpFLP, unlike in traditional RFLP.
Regions with core repeat sequences greater than 8 bp (base pairs) have been called minisattelite or long tandem repeat regions. Those with core repeat sequences of 2 to 8 bp are called microsattelite or short tandem repeat (STR). These STR are preferred because they are less susceptible to degradation. Automated STR analysis uses fluorescently labeled amplified product DNA. Real time PCR with multiple fluors is the latest technology in this area [7].
The short tandem repeats (STR) analysis, isolates DNA segments that all have the same sequence of repeating letters (ATCATCATC, for example). It organizes these repeating segments according to length, marks segments of a few different lengths, and then compares samples based on the presence or absence of same-length segments. Two samples that have as many as ten or twelve of these segments in common have very little chance — one in several million — of being from two different people [8].
Technique -Automated STR
The DNA is extracted from blood/ hair/ bones by digestion using enzymes and organic solvents. Subsequently to the extracted DNA multiple fluorescent dyes labelled probes and nucleotides are added. The dye is adhered to primer through NHS ester and amine linkage on the 5’ end of primer. The DNA is multiplied using multiplex PCR. Advantage of using the multiplex PCR is that at one time 13 different alleles can be labeled and multiplied simultaneously. The dye gives a specific colour tag to each PCR product. Based on the type of STR kit used (conventionally a 9 loci or 13 loci system is used); the probes are added [7]. 13 STR system has a standard set of loci in the human karyotype (Fig 1). The extracted and multiplied labeled DNA then undergoes the capillary electrophoresis step. (Fig 2 and 3) [9]. The argon laser analyses the DNA flowing through the capillary into a 13 wave front (Fig 4). When the two wave fronts of two different individuals are analyzed it can be seen clearly how two different human DNA differ when matched with each other at 11 different loci [8].
Advantages of STRs over traditional RFLP techniques
Discrete alleles from STR systems may be obtained due to their smaller size, which puts them in the size range where DNA fragments differing by a single base pair in size may be differentiated. In addition, smaller quantities of DNA, including degraded DNA, may be typed using STRs. STR processing is rapid and abundant STR are available in the human genome. Discrete alleles allow digital record of data that in turn allows the automated analysis through a computer.
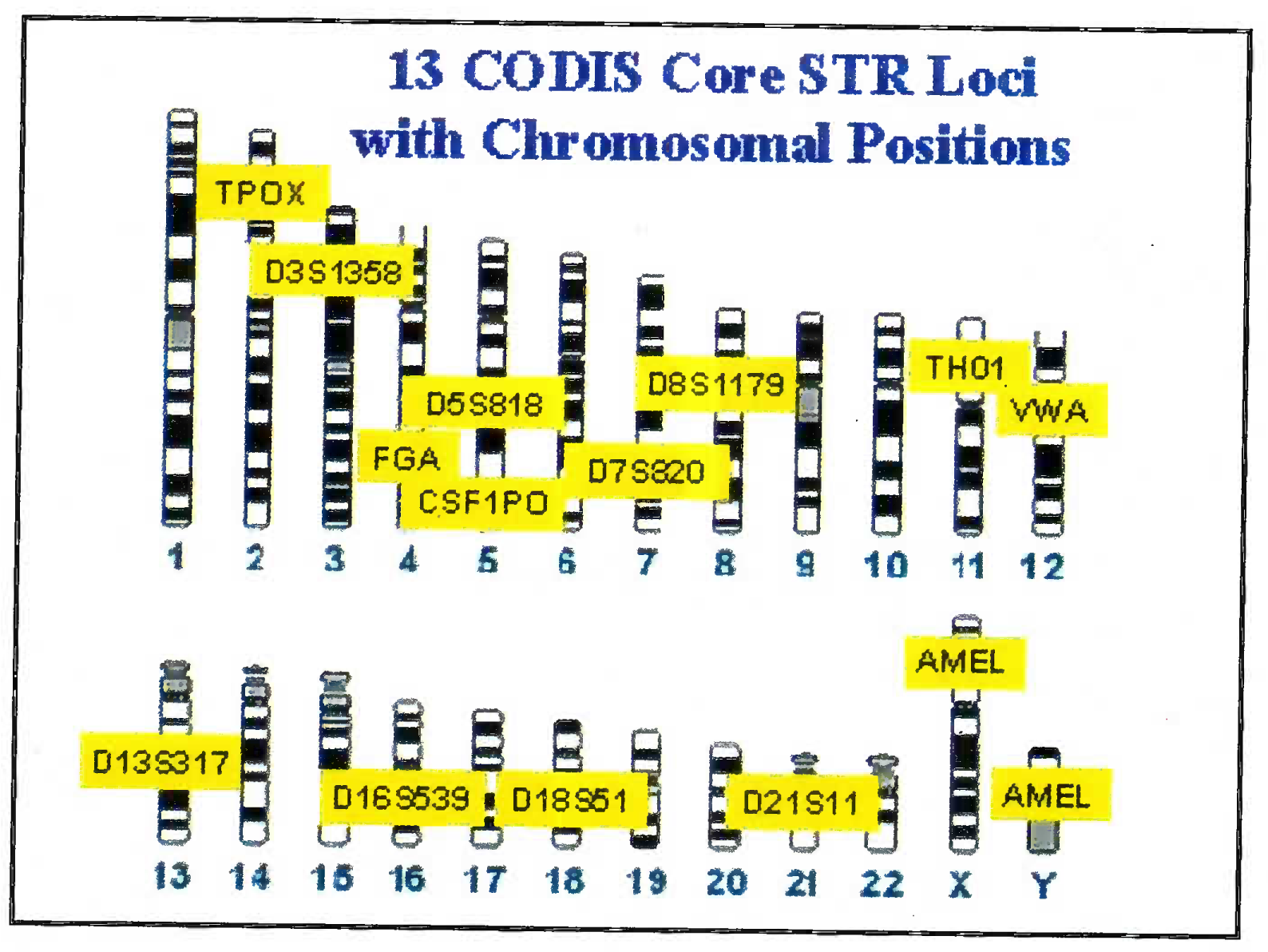
- 13 STR sites utilized in human chromosome in BNA analysis
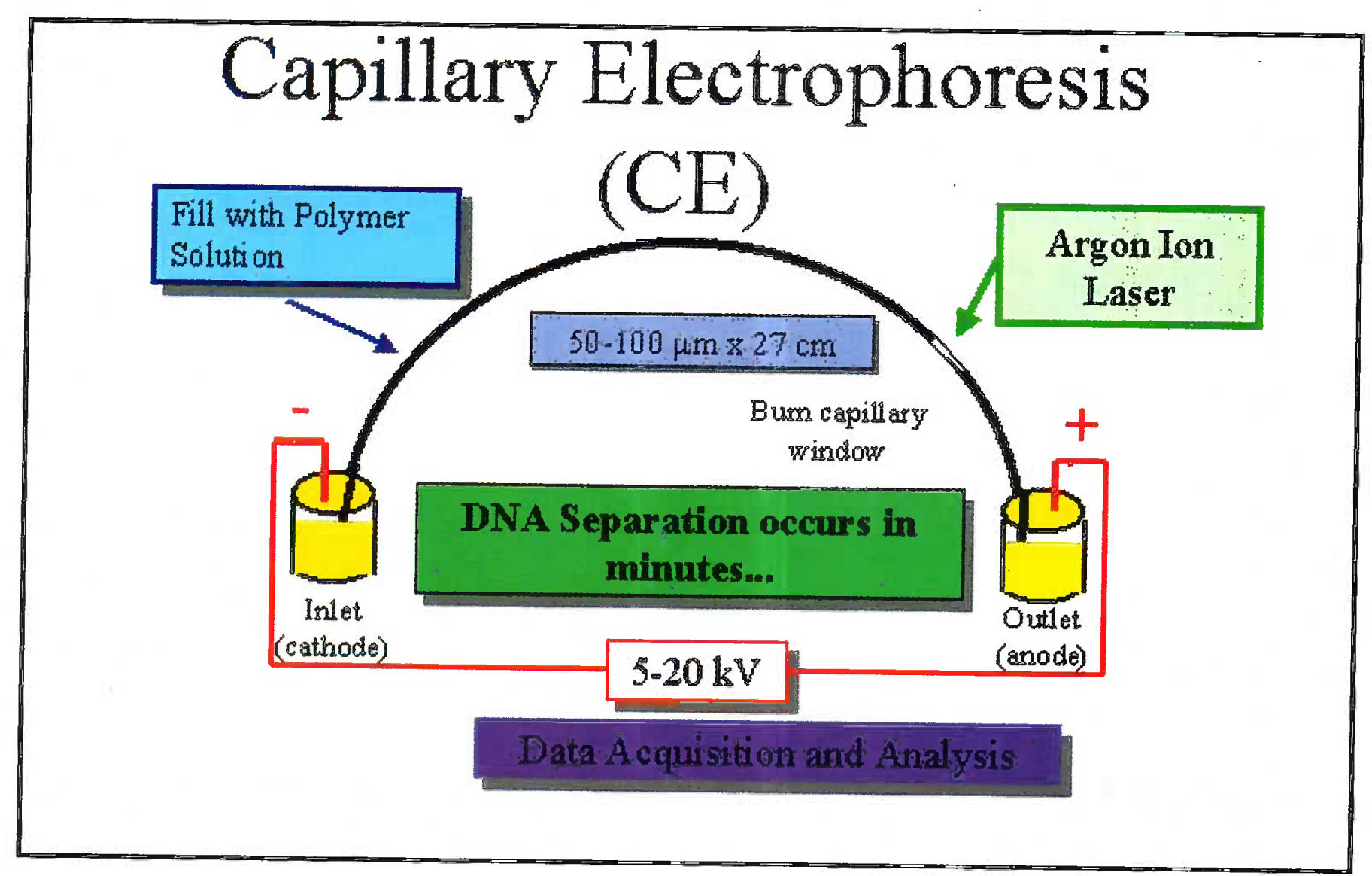
- Sketch diagram of electrophoresis where the extracted DNA flows through a capillary and the laser diffraction is analyzed
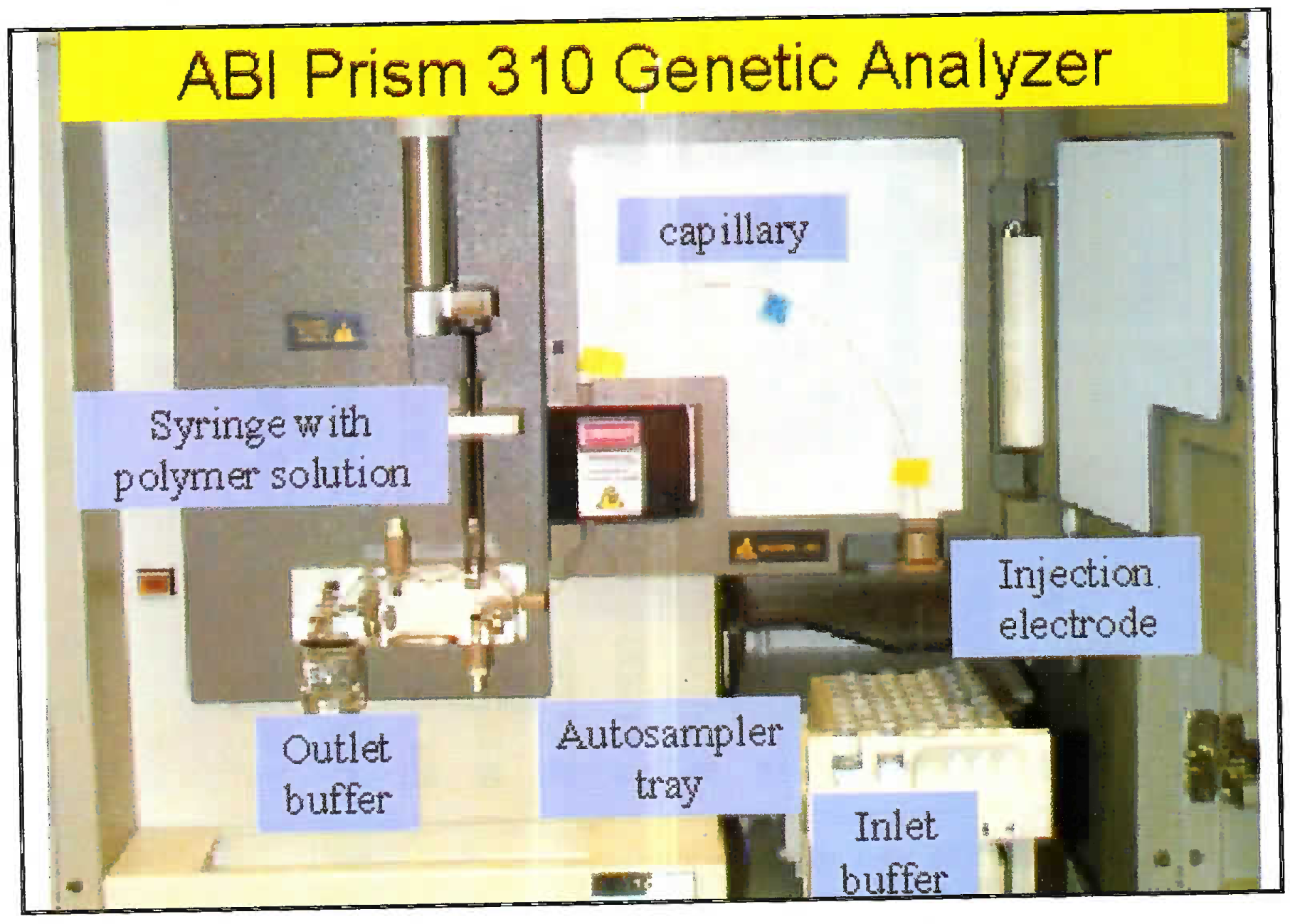
- Pictograph of capillary electrophoresis machine
- Wave front graph taken from electrophoresis machine showing the DNA of two different individuals differing at the selected 11 loci.
Mitochondrial DNA sequencing
Mitochondrial DNA (mtBNA) sequencing is a newer technology that has application in aviation pathology for two reasons. The mtBNA is present in numerous copies per cell and is applicable when nuclear DNA is extremely degraded as in air crashes where high temperatures degrade the nuclear DNA. The second reason is that mtBNA is 100% maternally inherited; therefore during identification of body the mitochondrial BNA of remains of the body can be easily compared with that of the mother or maternal uncles of the victim [10].
Mitochondrial DNA is a circular piece of DNA, 16,569 base pairs in length. Because no significant regions of repetitive DNA exist in mt DNA, only sequence polymorphism is typed. Unlike nuclear DNA, which is present in pairs of chromosomes, only a single sequence is present in the cell (homoplasmic) [11].
The region of mtDNA that is analyzed for human identification is the noncoding region known as displacement loop (D-loop) or control region. This loci spans 1100bp and contains two hypervariable regions (HV and HV1). The degree of polymorphism in the D-loop is so great that direct sequencing may be the most efficient method of typing mtDNA (Fig 5). The major three disadvantages of mtDNA analysis are (i) discriminatory power is 1:200 (ii) the procedure is expensive and (iii) heteroplasmy (few single base pair difference might be there in different cells of the same individual unlike the nuclear DNA) [12].
Steps (Figs 6 and 7) in mtDNA analysis (using human hair) include the following:-
Visual Analysis. The first step in the analysis of a hair involves a microscopic comparison of an evidentiary hair and a sample population of reference hairs. If the hair from a questioned source exhibits similar microscopic characteristics as hair from a known source, examiners perform mtDNA analysis to determine, on a molecular level.
Sample Preparation. Samples are cleaned prior to the mtDNA sequencing process to remove contaminating materials surrounding or adhering to the sample. The cleaning process for hair samples uses a detergent treatment in an ultrasonic water bath, which removes possible contaminating residues from the hair. The hair sample is then placed in an extraction solution and ground using a small mortar and pestle, resulting in a mixture that contains both the cellular material and the released DNA. For cleaning bone or tooth, an analyst sands the exterior to remove any extraneous material that may adhere to the surface.
DNA Processing. To extract DNA, analysts expose the cellular mixture from the sample preparation step to a mixture of organic or alkaline chemicals that separate the DNA from other biological material, such as proteins. Analysts then purify the DNA sample to prepare it for the amplification process, which makes many copies of the target DNA. After the DNA is copied, it is purified and quantified prior to determining the order of the bases in the DNA fragment. The sample undergoes a series of chemical reactions and is then placed in an instrument that “sequences” the bases in the DNA sample (Fig 7).
Data Analysis. There are established guidelines for the interpretation of differences and similarities between sequences. Basically, samples cannot be excluded as originating from the same source if sequence concordance (the presence of the same base or a common base at every position analyzed) exists between them. In cases of sequence concordance, at least two examiners independently analyze each sequence obtained [13].
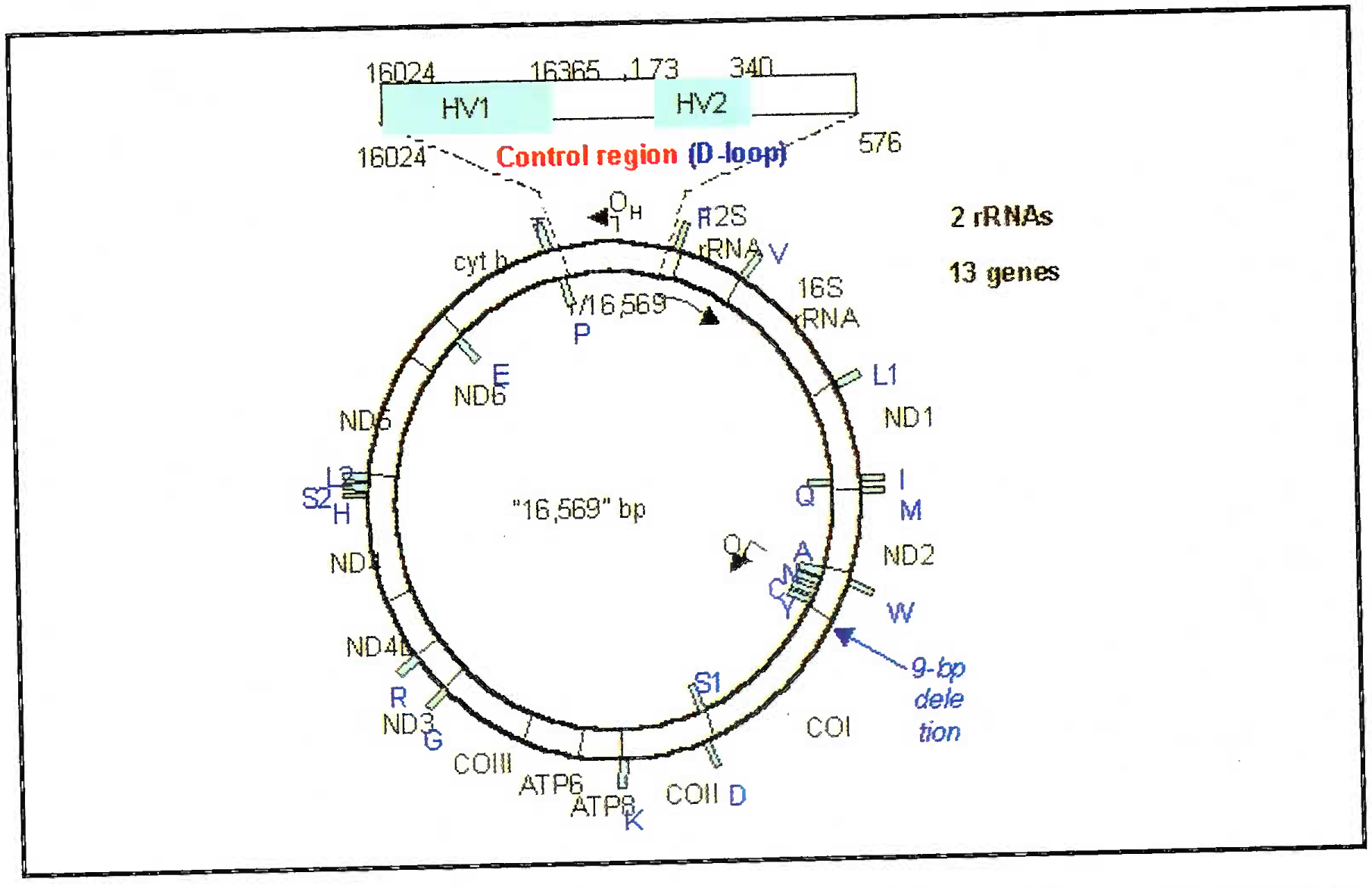
- Line diagram of mitochondrial DNA showing the D-loop and hypervariable areas
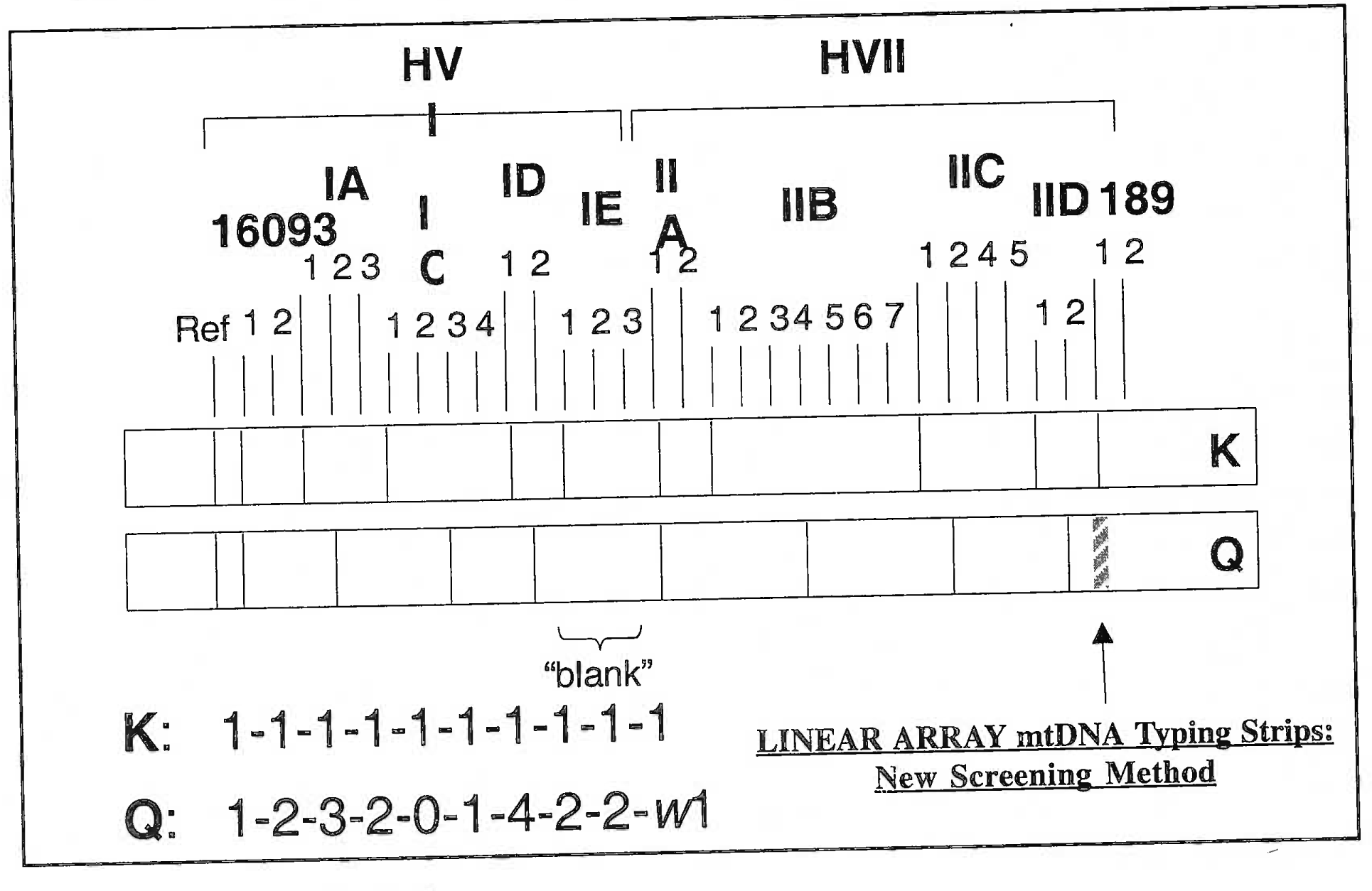
- Linear array of mitochondrial DNA on the strip of a known mtBNA (K) and mtBNA of question (Q). It is evident that there is a gross difference in the zone of bands formed by BNA’s of two different individuals
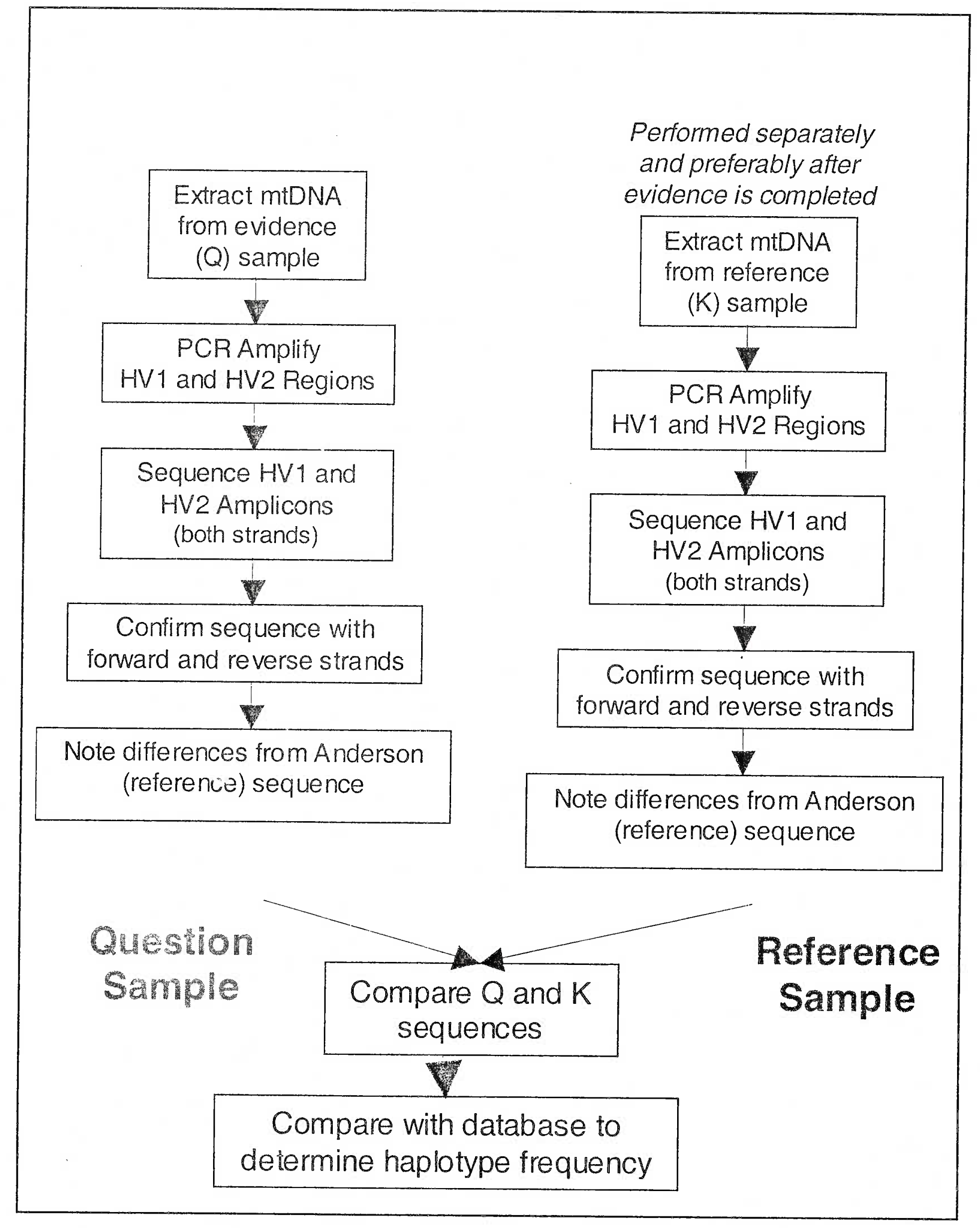
- Sequencing of Mitochondrial DNA at hypervariable region 1 of D-loop (Process for evaluation of mtDNA samples)
A software has also been innovated which provides indirect personality identification by establishment of biological kinship using a comparative analysis of nucleotide sequences (NS) of mtDNA in unidentified bodies and suspected relatives of mass disaster victims. The software supports databases with information on NS. To secure automatic search there are modes of cross processing. Results of identification are formed as lists of exclusions and tables with calculated frequencies of mitotypes of identification objects as well as protocols of a comparative analysis of nucleotide position in mtDNA sequences under comparison [14].
A bioinformatic tool was developed to assist with the victim identification initiative that followed the Swissair Flight 111 disaster. Making use of short tandem repeat (STR) DNA typing data generated with AmpFISTR Profiler Plus (PP) and AmpFISTR COfiler (CO) kits (Figs 8 and 9), the software systematically compared each available STR genotype with every other genotype. The matching algorithm was based on the search for (i) direct matches to genotypes derived from personal effects and (ii) potential kinship associations between victims and next-of-kin, as measured by allele sharing at individual loci [15].
Quality assurance and standards
Quality assurance issues and concerns are important to any service laboratory testing and have been an area of emphasis in DNA testing. Inter laboratory comparisons have consistently demonstrated the great precision of RFLP and PCR testing of DNA. Proficiency surveys report a good DNA laboratory should have mean fragment size measurements that differ only by a few bases and the range of results should be well within plus or minus 2.5% matching window used by most DNA laboratories [16]. The resolution of STR system should be between 1 and 300bp so that microvariant alleles can be picked up. In mt DNA analysis, in instances, few copies of the DNA of interest might be present (if a sample is degraded), the presence of any foreign DNA in the sample can harm the analysis. Any extraneous DNA that is amplified with the evidence has the potential to interfere with the interpretation of the sequence data. The laboratory should follow general and specific precautions designed to minimize contamination, including the physical separation of pre-and post-amplification areas; the separation of evidence from questioned and known sources in time and often, space; the proper cleansing of work spaces and instruments; and the use of control samples. The laboratory should use positive and negative controls in mtDNA processing to monitor amplification and sequencing and should not proceed with the mtDNA analysis if these controls fail to meet established criteria [17].
Specimen collection
DNA analysis can be performed on a wide variety of specimens.
(a) Whole Blood-EDTA or CPD anti coagulated blood remains the specimen of choice for DNA laboratories. Heparin is not recommended. However as blood is first to putrefy it is generally haemolysed or contaminated by bacteria.
(b) Tissue-Soft tissue DNA like that of liver is first to degrade. The bone and teeth are most stable sources and perhaps for this reason informative mitochondrial DNA is routinely obtained from skeletal remains, which may be decades old. Great care should be taken to prevent contamination of the specimen by other sources of DNA. Specimen should be collected using gloves and sterile instruments. The specimens should be kept cold or preferably frozen. Desiccation may be an adequate method of storage of some DNA specimens like blood/ bone. Tissues in formalin are not optimal but can be used for PCR-based DNA testing [18].

- Amp FISTR system: 9 STR system showing the DNA of individual separated on known loci using three fluorescent labels. The red row is the standard molecular ladder with which the PCR product size can be compared
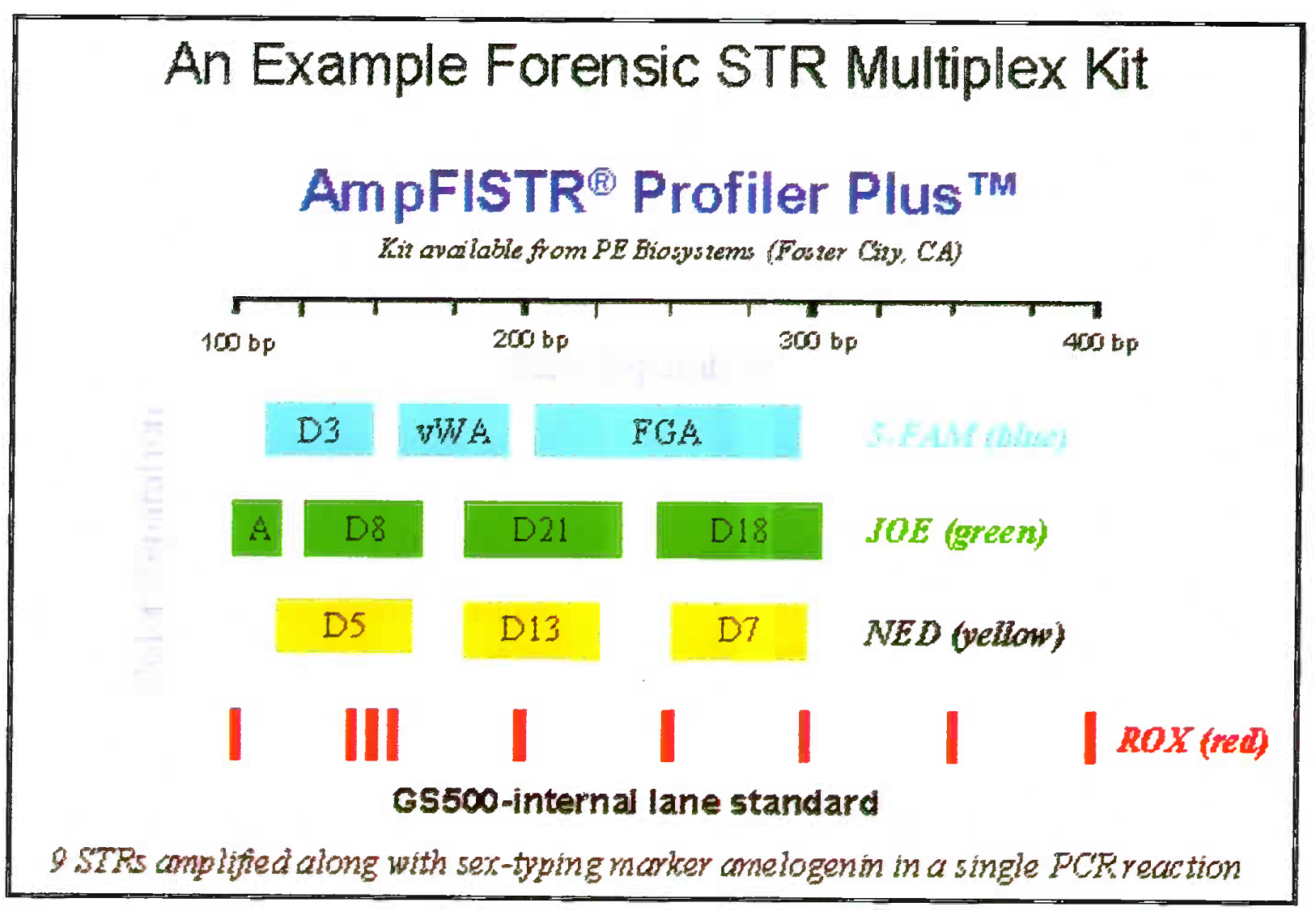
- 9 STRs amplified along with sex-typing marker amelogenim in a single PCR reaction
Conclusion
The problem of identification of human remains is still one of the most difficult and pressing problems of aviation pathology. Such identification is particularly difficult in cases of intense destruction caused by exposure to physical factors (explosion, contusion, fire, compression, etc.). Approaches to identification in such cases are less available and differ essentially from traditional approaches used, for example, in cases when the remains are exposed to natural long-acting factors. The DNA-typing identification modality has proved to be a valuable component of the large arsenal of identification tools deployed in the aftermath of mass disaster. The future of all DNA analyses looks promising as techniques for smaller-scale and higher-output testing become available. While mtDNA analyses do not provide the discrimination potential of some nuclear DNA tests, mtDNA data often are the only information that examiners can gather from degraded evidence, which is either old or has been exposed to the environment for a significant period of time. The development of forensic mtDNA sequencing over the past decade has been helpful to many past cases and will continue to provide useful information to the law enforcement community in the future.
Recommendations
This paper provides an overview of the utility of DNA testing in the identification of aircrash victims. Establishment of a DNA repository to store reference blood samples to facilitate the identification of Indian Air Force aircrew remains may be worthwhile. The DNA repository can be developed in form of small blood stained card with preservative for genomic DNA and few strands of plucked hair for mitochondrial DNA during commissioning or enrollment into Indian Air Force.
References
- Possible express identification of human remains in case of their intense destruction after exposure to physical factors. Sud Med Ekspert. 2002;45(4):35-9.
- [Google Scholar]
- Practical use of the molecular-genetic technologies in solving the tasks of forensic medical identification of remains in emergencies with huge human death toll. Sud Med Ekspert. 2004;47(5):31-40.
- [Google Scholar]
- Medico-legal investigations of the Airbus, A320 crash upon Mount Ste-Odile, France. J Forensic Sci. 1994;39(5):1147-52.
- [Google Scholar]
- Disaster victim identification of military aircrew, 1945-2002. Aviat Space Environ Med. 2003;74(11):1198-200.
- [Google Scholar]
- Forensic Identity testing by DNA analysis In: Henry JB, ed. Clinical Diagnosis and management by Laboratory Methods (20th ed). Philadelphia: WB Saunders Company; 2002. p. :1583-1623. In:
- [Google Scholar]
- Characterisation of new mini STR loci to aid analysis of degraded DNA. J Forensic Sci. 2005;50:43-53.
- [Google Scholar]
- Genetics and genomics of Core STR loci used in human identity testing. J Forensic Sci. 2006;51(2):253-65.
- [Google Scholar]
- Forensic DNA tying by capillary electrophoresis using ABI prism 310 genetic analyzer for STR analysis. Electrophoresis. 2004;25:1397-1412.
- [Google Scholar]
- Application of mtDNA sequence analysis in forensic casework for the identification of human remains. Forensic Sei Int. 2000;113(1-3):103-7.
- [Google Scholar]
- A duplex real-time qPCR assay for the quantification of human nuclear and mitochondrial DNA in forensic samples: Implications for quantifying DNA in degraded samples. J Forensic Sci. 2005;50(5):1044-60.
- [Google Scholar]
- A simplified method for mitochondrial DNA extraction from head hair shafts. J Forensic Sci. 2005;50(5):1119-22.
- [Google Scholar]
- Isenberg Forensic mitochondrial DNA analysis: A different crime-solving tool. Forensic Science Communications. 1999;54:202-324.
- [Google Scholar]
- Design of the automatic system of processing data on mitochondrial DNA “mtDNAbase” sequestration for expert identification of the unidentified bodies under the conditions of mass reception of human remains. Sud Med Ekspert. 2002;45(5):15-21.
- [Google Scholar]
- Enhanced kinship analysis and STR-based DNA typing for human identification in mass fatality incidents: The Swissair flight 111 disaster. J Forensic Sci. 2004;49(5):939-53.
- [Google Scholar]
- Inter laboratory comparisons of autoradiographic DNA profiling measurements. Anal Chem. 1994;66:3303-09.
- [Google Scholar]
- Quality control of PGR primers used in multiple STR amplification. Forensic Science Int. 2001;119:87-96.
- [Google Scholar]
- Recovery and stability of DNA in samples of forensic science significance. Forensic Sci Rev. 1992;4:67-70.
- [Google Scholar]





